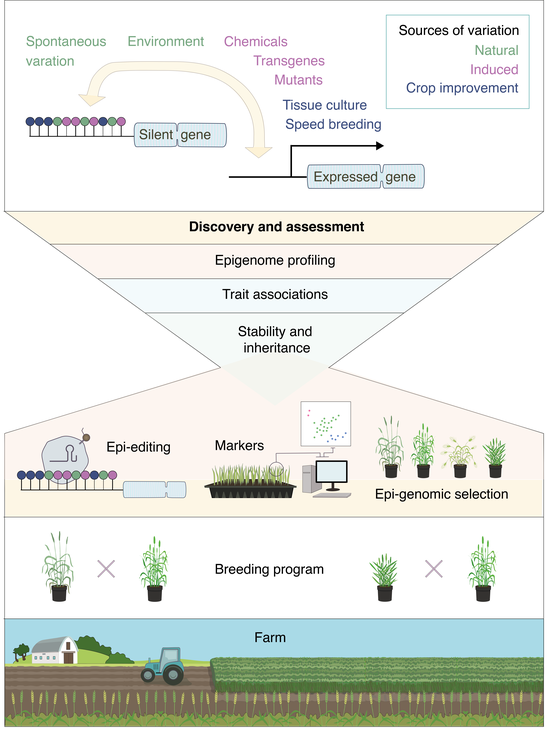
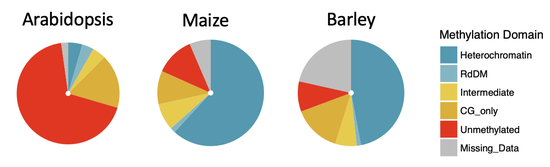
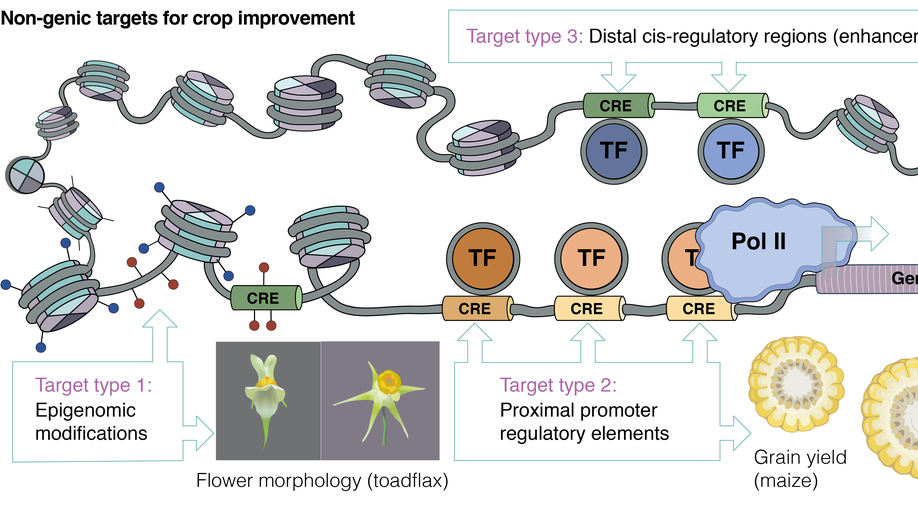
Recent News

PhD position available on crop epigenomics and enhancers
We have a funded PhD scholarship opportunity in the lab. Check out the details in this post or get in contact for more information. The Scholarship is officially listed on the UQ Graduate School page search for Peter Crisp. The position is a fully funded PhD scholarship for a domestic or international student (stipend + tuition + health cover). More details on the scholarship scheme can be found here on the UQ Graduate School Earmarked Scholarship page.

The Lab Opened
There has been a lot going on lately… including the grand opening of the Lab. We are here! Excited to be at UQ. Located in the John Hines Building, St Lucia.
Lab Opening Feb 2020
The lab will be opening at the University of Queensland, St Lucia Campus, in the School of Agriculture and Food Sciences beginning Feb 2020. We will be looking for students! Contact me for more information.